The goal of paleoTS is to allow the user to simulate and fit time-series models commonly used to understand trait evolution in paleontology. Models include random walks, stasis, directional trends, OU, covariate-tracking, punctuations and more. Model fitting is done via maximum likelihood.
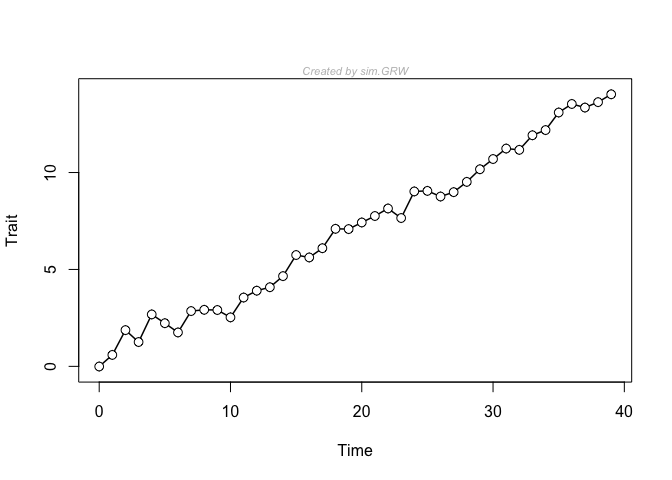
This is a simple example in which a time-series is generated, plotted, and then fit with three common models in paleobiology. The generating model is a general (also called biased) random walk, with a pretty strong trend parameter. Usually, this model receives just about all of the available model support with these generating parameters.
library(paleoTS)
y <- sim.GRW(ns = 40, ms = 0.3)
plot(y)
fit3models(y)
#>
#> Comparing 3 models [n = 40, method = Joint]
#>
#> logL K AICc dAICc Akaike.wt
#> GRW -26.86719 3 60.40106 0.00000 1
#> URW -37.85943 2 80.04318 19.64213 0
#> Stasis -113.33758 2 230.99949 170.59844 0Take a look at the vignette “paleoTS_basics” for more of an introduction to this package.
paleoTS should be installed from CRAN.